A new study by Miguel Gonzalez-Lozano (MCN) and colleagues demonstrates how cross-linking mass spectrometry can contribute to the discovery of novel synaptic protein interactions. The study was published in Science Advances on February 19, 2020.
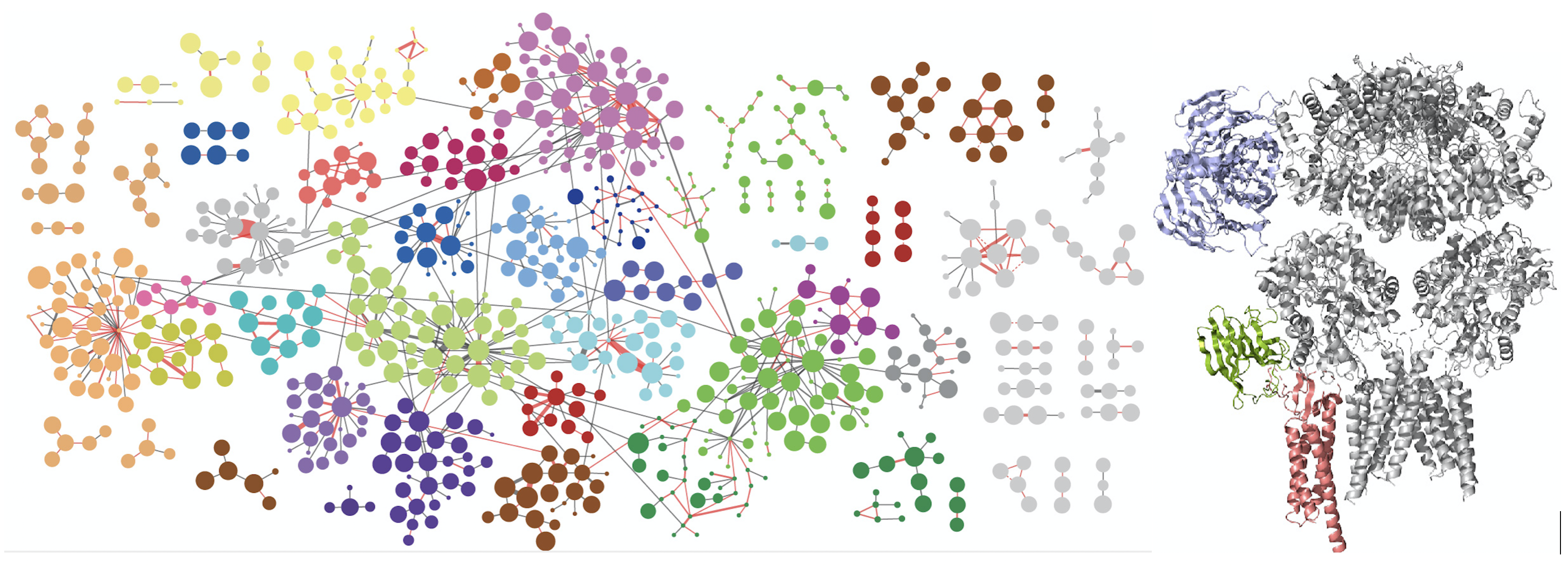
Synaptic transmission requires functionally specialized molecular machineries constituted by a multitude of interacting synaptic proteins. In this study we made use of recent advances in cross-linking mass spectrometry (XL-MS) in combination with biochemical and computational approaches to reveal the architecture and assembly of synaptic protein complexes from mouse brain hippocampus and cerebellum. The 12.000 unique lysine-lysine cross-links comprise connections within and between 2,362 proteins. This extensive collection was the basis to identify novel protein partners, to model protein conformational dynamics and to delineate within and between protein interactions of main synaptic constituents, such as Camk2, the AMPA-type glutamate receptor and associated proteins. Using various biochemical and computational approaches, we validated the fidelity of our procedure and investigated protein interactions and dynamics for key synaptic proteins, such as Camk2 and the AMPA receptor. This extensive resource provides a novel perspective on protein structures, assemblies and interactions of the synaptic proteome. The protein interaction resource that we made is easily accessible via a web-based platform to provide new entries into exploration of all protein interactions. The paper can be found here
Left figure: The network of interacting synaptic proteins as visualized through crosslinking mass spectrometry. Right figure: three interacting proteins (colored) of the AMPA receptor positioned onto the AMPAR 3D structure based on the crosslinking data.