Our goal is to understand the specific molecular, cellular and network adaptations that occur in the brain in the earliest stages of Alzheimer’s diseasse, and to test how these adaptations can be used for early disease diagnosis and intervention.
Research focus
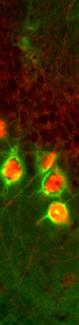
My research currently focusses on
1. Identification of additional interneuron subtypes whose dysfunction may contribute to early AD pathogenesis;
2. The impact of interneuron hyperexcitability on neuronal networks in vivo, and how to translate these findings to earlier and better diagnosis of AD in humans;
3. The identification of specific channels and receptors that contribute to interneuron hyperexcitability and may be targeted for pharmacological intervention;
4. The specific role of interneuron hyperexcitability in the formation and reactivation memory engrams;
5. The role of interneuron hyperexcitability in mouse models of sporadic AD, such as hAPOE3/4 mice.
Key publications
Reducing Hippocampal Extracellular Matrix Reverses Early Memory Deficits in a Mouse Model of Alzheimer’s Disease.
MJ Vegh, CM Heldring, W Kamphuis, S Hijazi, AJ Timmerman, KW Li, P van Nierop, HD Mansvelder, EM Hol, AB Smit, and RE van Kesteren, Acta Neuropathol Comm 2:76 (2014)
Early Restoration of Parvalbumin Interneuron Activity Prevents Memory Loss and Network Hyperexcitability in a Mouse Model of Alzheimer’s Disease.
S Hijazi, TS Heistek, P Scheltens, U Neumann, DR Shimshek, HD Mansvelder, AB Smit, and RE van Kesteren, Mol. Psychiatry doi:10.1038/s41380-019-0483-4 (2019)
Hyperexcitable Pv Interneurons Render Hippocampal Circuitry Vulnerable to Amyloid Beta.
S Hijazi, TS Heistek, R van der Loo, HD Mansvelder, AB Smit, and RE van Kesteren, iScience doi:10.1016/j.isci.2020.101271 (2020)
Technology
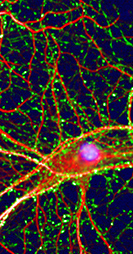
For an overview of our facilities see High-Content Screening at CNCR.
For examples of our work involving high-content screening see
High Content Screening in Neurodegenerative Diseases.
S Jain, RE van Kesteren, and P Heutink, J Vis Exp e3452 (2012)
Combined Cellomics and Proteomics Analysis Reveals Shared Neuronal Morphology and Molecular Pathway Phenotypes for Multiple Schizophrenia Risk Genes.
M Rosato, S Stringer, T Gebuis, I Paliukhovich, KW Li, D Posthuma, PF Sullivan, AB Smit, and RE van Kesteren, Mol. Psychiatry doi:10.1038/s41380-019-0436-y (2019)